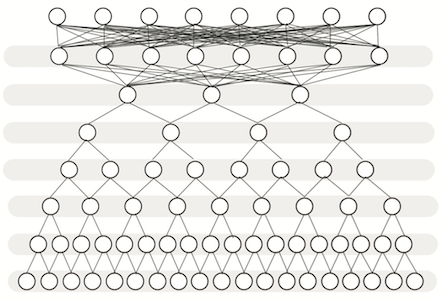
Deep learning expression model
ExPecto leverages deep learning-based sequence models trained on chromatin profiling data, and integrated with spatial transformation and regularized linear models. This framework does not use any variant information, enabling prediction of the expression effect for any variant, even those that are rare or have never been previously observed.
Cell-type-specific expression
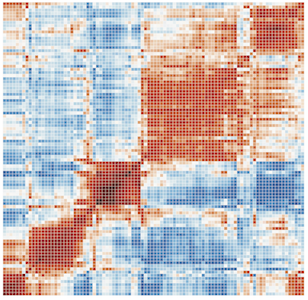
ExPecto models make highly accurate cell-type-specific predictions of expression solely from DNA sequence. With ExPecto, the tissue-specific impact of human gene transcriptional dysregulation can be systematically probed 'in silico' - at a scale not yet possible experimentally.
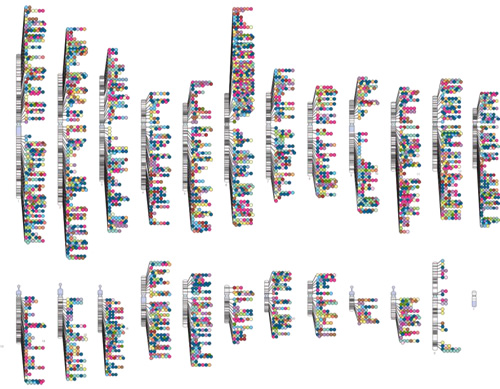
Disease-risk variants
ExPecto can prioritize putative causal variants associated with human traits and diseases, and predict new disease impact mutations. Novel ExPecto predictions of causal variants for Crohn's disease, ulcerative colitis, Behcet's disease and HBV infection have all been experimentally validated, demonstrating the power of ExPecto prioritization over traditional GWAS studies.